 |
|
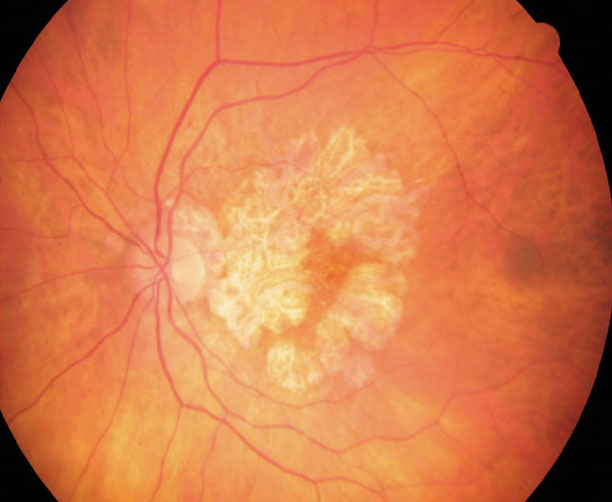
RPE cells from GA patients show constitutive differences in visual cycle processes and melanosome function. Photo: Anna Bedwell, OD. Click image to enlarge. |
Innovative studies are needed to model geography atrophy and prevent or delay its progression. Researchers in Australia differentiated induced pluripotent stem cells generated from patients with geographic atrophy (GA) and healthy individuals to retinal pigment epithelium (RPE) cells and then identified important constitutive differences in RPE homeostasis associated with GA when compared with healthy RPE cells.1
Transcriptomics and proteomics approaches identified molecular pathways significantly upregulated in GA, including in mitochondrial functions, metabolic pathways and extracellular cellular matrix reorganization. The study identified five significant protein quantitative trait loci that regulate protein expression in the RPE and in GA—two of which share variants with cis- expression quantitative trait loci, including proteins involved in mitochondrial biology and neurodegeneration. Investigating mitochondrial metabolism confirmed that mitochondrial dysfunction as a core constitutive difference in the RPE cells from patients with GA. Retinoid and pigmentation-related proteins were amongst the most significantly differentially expressed between the control and GA samples.
“Taken together, our data suggest that RPE cells from the GA cohort show constitutive differences in visual cycle processes and melanosome function to healthy RPE cells,” the researchers noted in their paper in Nature Communications. “Whether these differences are simply associated or causative of phenotypes remains to be assessed.”1
Speaking to Genetic Engineering and Biotechnology News, the researchers noted that the discovery of new genetic signatures of the disease will lead to better diagnosis and treatment of age-related macular degeneration before progressing to GA. The molecular signatures could potentially be used for treatment screenings using patient-specific cells in a dish. The team continues to build a program of research interested in stem cell studies to model disease at very large scale to do screening for future clinical trials.2
“At the smallest scale we’ve narrowed down specific types of cells to pinpoint the genetic markers of this disease,” they said. “This is the basis of precision medicine, where we can then look at what therapeutics might be most effective for a person’s genetic profile of disease.”
1. Senabouth A, Daniszewski M Lidgerwood GE, et al. Transcriptomic and proteomic retinal pigment epithelium signatures of age-related macular degeneration. Nat Commun. July 26, 2022. [Epub ahead of print]. 2. Macular degeneration molecular signature may aid diagnosis, treatment. Genetic Engineering and Biotechnology News. July 26, 2022. www.genengnews.com/eye-disorders/macular-degeneration-molecular-signature-may-aid-diagnosis-treatment/. Accessed August 4, 2022. |

